The 2015-2016 flu season is expected to peak this month. Though this year is expected to be comparatively mild, annual flu season claims 36,000 lives and leads to millions of hospitalizations in the United States. A flu pandemic can result in a more catastrophic impact, as witnessed by the 1918 Spanish flu and the recent 2009 swine flu. We spoke with Jun Wang, University of Arizona College of Pharmacy, about his research on the M2 proton channel of influenza A viruses.
What is the connection between your research and influenza?
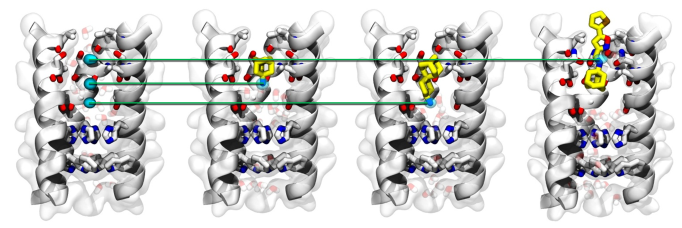
M2 proton channel is universally expressed in the viral membranes of all influenza A viruses. It is a multifunctional protein that is absolutely essential for the viral replication. Among the 97 residues, the transmembrane domain (25-46) forms a homo-tetrameric four-helix bundle which mediates selective proton conductance. This function is essential for the viral uncoating once the virus is engulfed in the endosome. M2 is the known drug target of amantadine. However, more than 95% of current circulating influenza A viruses carry mutated M2 channels which render them resistant to amantadine, among which S31N is the predominant mutant. Given the relevance of M2 as an antiviral drug target, we are interested in understanding the mechanism of amantadine in inhibiting the wild-type M2 channel. Once we are convinced we understand this process, we would like to apply our knowledge to a practical exercise which is to design novel channel blockers targeting the S31N mutant.
Why is your research important to those concerned about influenza?
Annual flu season claims 36,000 lives and leads to millions of hospitalizations in the United States. Flu pandemic results in more catastrophic impact as witnessed by the 1918 Spanish flu and the recent 2009 swine flu. However we are limited in countermeasures in prophylaxis and treatment of flu infection: only one oral drug, Tamiflu, is still in use. Given the lessons we learned from antibiotics and antivirals, there is no doubt that with the increasing prescription of Tamiflu it is only a matter of time before a majority of the sensitive viruses will evolve to become resistant to it. The shocking reality is that a large number of Tamiflu-resistant strains have already been identified from human patients. Thus, there is a clear need for the next generation of novel antivirals. The S31N inhibitors we discovered represent the second line of defense should Tamiflu fail to confine an influenza A virus outbreak during the next flu pandemic. S31N inhibitors have been shown to be highly potent in inhibiting multidrug-resistant influenza A strains and have synergistic antiviral effect with Tamiflu. Thus, they can be used in combination with Tamiflu to decrease the pace of resistance evolution.
How did you get into this area of research?
I began studying the M2 proton channel as a graduate student at the University of Pennsylvania in the lab of Dr. William F. DeGrado. The DeGrado lab has a long standing reputation in de novo design of four-helix bundles with novel functions. As M2 is a natural four-helix bundle with profound proton selectivity; the DeGrado lab was interested in understanding the structure and function relationship of M2 as well as the drug inhibition mechanism of M2. For example, how conformational change is coupled with proton conductance?; why M2 selectively conducts proton in a unidirectional manner?; and how does amantadine block the M2 channel? The knowledge gathered from such studies are critical as they serve as invaluable guides not only for advancing our fundamental understanding, but also the design of novel channel blockers. I first started by addressing the question regarding where the pharmacologically relevant drug binding site is for amantadine in M2. This part of the work was done in close collaboration with Dr. Mei Hong, who is now a professor in chemistry at MIT. Other major contributors in the DeGrado lab working on this project include Dr. Rudresh Acharya, an assistant professor at the National Institute of Science Education and Research in India, and Dr. Yibing Wu, a senior specialist in the DeGrado Lab.
How long have you been working on it?
I have been working on the M2 proton channels for 10 years since I began my graduate research in 2006. I continue working on this target since I became a PI at the University of Arizona. The primary focus of my laboratory in this project is to further advance S31N inhibitors to the stage of filling an Investigational New Drug application. The DeGrado lab continues working on the biophysical aspects of M2.
Do you receive public funding for this work? If so, from what agency?
The drug discovery of M2-S31N inhibitors are funded by both the NIAID, NIH (AI119187) and the PhRMA foundation 2015 Research Starter Grant in Pharmacology and Toxicology. We are also particularly grateful to NIGMS for their support of DeGrado’s work on M2 through GM056423.
Have you had any surprise findings thus far?
M2-S31N was traditionally tagged as an undruggable target because decades of traditional medicinal chemistry campaign failed to yield a hit compound. Thus, the discovery of the first S31N inhibitor by itself was a surprise finding. With this tool compound in hand, we were able to solve the solution NMR structure of S31N mutant in the drug-bound form. The structure revealed that S31N inhibitor binds to the mutant channel in a reverted orientation compared with that of amantadine in wild-type M2. The drug-bound S31N structure represents the Openout-Closein conformation which was not captured by previous structures.
What is particularly interesting about the work from the perspective of other researchers?
First, we resolved the controversy regarding the pharmacologically relevant drug binding site of M2, which allows other researchers to focus their efforts on the more relevant channel pore for their drug design. Second, using molecular dynamics simulations, we identified three hot spots aligning along the channel pore where the positive charged ammonium from the M2 channel blockers bind to. This is a reminiscent of how potassium channel blockers work, although the detailed mechanisms are obviously very different. This mechanism can be applied to guide the design of inhibitors targeting other ion channels.
What is particularly interesting about the work from the perspective of the public?
From the public perspective, the S31N inhibitors we discovered offer an opportunity for the urgently needed next generation of antivirals. As S31N inhibitors have no overlapping drug resistance profile with Tamiflu, they can be used either alone to treat infections with Tamiflu-resistant virus or used in combination with Tamiflu to achieve better therapeutic outcome. Moreover as M2-S31N is prevalent among circulating influenza A strains, S31N inhibitors are expected to have broad-spectrum antiviral activity.